Munging annotations from Praat TextGrid files with crowsetta to fit a first-order Markov model of birdsong syntax#
A common need when working with annotations for animal vocalizations is to transform them to perform a certain analysis. This is a sort of data munging. In this vignette, we provide a simple example of munging annotations to build a model of birdsong syntax.
We will work with a dataset of Bengalese finch song annotated in a format used by “evsonganaly”, a MATLAB GUI for annotating song. We want to build a matrix of the transition probabilities between syllables in the song, that is, a first-order Markov model.
Download this page as a Jupyter notebook!
To work with this tutorial interactively, we suggest downloading this notebook! Click on the download icon in the upper right to download a Markdown file (with ‘.md’ extension) that you can run as a Jupyter notebook after installing Jupyter lab and jupytext.
Workflow#
Here’s the steps we’ll follow:
Write code for analysis
Load .not.mat files saved by evsonganaly with crowsetta, and convert to crowsetta Annotations with Sequences
Munge data
Compute transition matrices
For this vignette, we use annotations from the Bengalese finch song repository, by Nicholson, Queen, Sober 2017 [1], adapted under CC BY 4.0 License.
0. Write code for analysis#
Before we can analyze our data, we need to know what analysis we’re doing. It can save you a lot of time to write a rough draft of this analysis code first, and test it with some toy data that you generate.
We also do this first to help understand what format we’re going to need to get our data into below.
Our goal here is to fit a first-order Markov model of the transition probabilities in the song of Bengalese finches. This model has been used in several previous studies[2][3], although it was later shown[4] that such a model does not completely describe the statistics of the song. In spite of that, a first-order Markov model is very convenient to fit, and can give us a useful first-order approximation (literally!) of singing behavior.
To build such a model for Bengalese finch song, we consider the song syllables to be states. We construct a transition matrix \(P\) where the rows are the state we’re transitioning from, the columns are the state we’re transitioning to, and thus the value in each cell \(p_{i, j}\) is the probability of transitioning from state \(i\) to state \(j\). We construct this matrix given a set of sequences \(X\), where each sequence \(x_t\) is a chain of states from a set \(S\), which in our case will be the set of syllable labels. To construct the matrix, we count the number of occurrences of each of the possible pairwise transitions from one state to another. Then, to compute the probabilities, we divide the counts for each pair by the total number of transitions. To replicate the approach used in [3], we will add start and end states to our transition matrix, that correspond to the start and end of each song bout, instead of computing a separate vector of initial state probabilities \(P_{init}\).
We will write a “pipeline” to compute this, which has basically two steps.
Using annotations loaded into crowsetta, get the unique set of states, and the counts of every occurrence of transitioning from one state to another.
Use the states and counts to compute the probabilities for the first-order Markov model
We write functions for step 1 and 2 below.
The functions are written in such a way that,
after cleaning and transforming our loaded annotations,
we pass them into the second function, and it will call the first one for us
when it computes the transition matrix.
Note
This code is adapted from yardencsGitHub/tweetynet under BSD license.
As used for the paper Cohen, Y., Nicholson, D., Sanchioni, A., Mallaber, E., Skidanova, V., & Gardner, T. (2022). Automated annotation of birdsong with a neural network that segments spectrograms. eLife. https://doi.org/10.7554/eLife.63853
"""functions for analyzing sequences of Bengalese finch syllable labels"""
from __future__ import annotations
from collections import Counter, namedtuple
import crowsetta
import numpy as np
def states_and_counts_from_labels(labels: list[np.ndarray],
start_state:str = '@',
end_state:str = '!') -> set:
"""Get ``states`` and ``counts``
from a list of numpy arrays of string characters,
to build a transition matrix
showing the probability of going from
one state to another.
Parameters
----------
labels : list
Of numpy.ndarray of string labels,
taken from the ``Sequence.label``
attributes of ``crowsetta.Annotation`` instances.
Will be used to count transitions
from one label to another
in the sequences of syllables.
start_state : str
Single character used to indicate the
start of a bout of song. Default is '@'.
end_state : str
Single character used to indicate the
end of a bout of song. Default is '!'.
Returns
-------
states : set
of characters, set of
unique labels
that occur in the list of
``Annotation``s.
counts : collections.Counter
where keys are transitions
of the form "from label,
to label", e.g. "ab",
and values are number of
occurrences of that transition.
Notes
-----
Note that characters for start and end states
are appended to the beginning and end of
each string in ``labels`` to build the
transition matrix as in Jin Kozhevnikov 2011.
"""
# make sequences with start and end states
seqs = []
for seq_labels in labels:
seq_labels = np.concatenate((np.array([start_state]), seq_labels, np.array([end_state])))
seqs.append(seq_labels)
# set of (unique) states
states = sorted(
set([state
for seq in seqs
for state in seq
])
)
counts = Counter()
for seq in seqs:
transitions = zip(seq[:-1], seq[1:])
for transition in transitions:
counts[transition] += 1
return states, counts
def row_norm(mat: np.ndarray) -> np.ndarray:
"""Normalize matrix rows, so they sum to one"""
return mat / mat.sum(axis=1)[:, np.newaxis]
TransitionMatrix = namedtuple('TransitionMatrix',
field_names=('counts',
'matrix',
'states'))
def transmat_from_labels(labels: list[np.ndarray],
thresh: float | None = None,
start_state:str = '@',
end_state:str = '!') -> TransitionMatrix:
"""From list of labels from ``crowsetta.Annotation``s,
returns ``TransitionMatrix`` tuple with fields
'counts', 'matrix', and 'states'
Parameters
----------
labels : list
Of numpy.ndarray of string labels,
taken from the ``Sequence.label``
attributes of ``crowsetta.Annotation`` instances.
Will be used to count transitions
from one label to another
in the sequences of syllables.
thresh : float
threshold used to smooth probabilities.
If not None, any probabilities less
than threshold are set to 0.0 and then
the transition matrix is again row
normalized
start_state : str
Single character used to indicate the
start of a bout of song. Default is '@'.
end_state : str
Single character used to indicate the
end of a bout of song. Default is '!'.
Returns
-------
trans_mat : TransitionMatrix
A NamedTuple with fields 'counts', 'matrix', and 'states'.
Notes
-----
Note that characters for start and end states
are appended to the beginning and end of
each string in ``labels`` to build the
transition matrix as in Jin Kozhevnikov 2011.
The returned matrix will have (:math:`s - 1` rows
and :math:`s` columns, where :math`s` is the
number of states, (i.e., ``len(trans_mat.states)``).
This is because there is no probability
of transitioning from the end state to any other.
"""
states, counts = states_and_counts_from_labels(labels, start_state=start_state, end_state=end_state)
counts_arr = np.array(
[[counts[i, j] for j in states] for i in states], dtype=float
)
# remove row for end state since probability of
# transitioning *from* that state to any other is zero.
# we use a mask to keep only rows we want
end_state_ind = states.index(end_state)
rows = np.ones(counts_arr.shape[0], dtype=bool)
rows[end_state_ind] = False
counts_arr = counts_arr[rows]
trans_mat = row_norm(counts_arr)
if thresh is not None:
trans_mat[trans_mat < thresh] = 0.0
trans_mat = row_norm(trans_mat)
return TransitionMatrix(counts=counts, matrix=trans_mat, states=states)
1. Load data with crowsetta#
We change directories so we can access the ./data directory we have set up for our project, following good practices.
cd ..
/home/docs/checkouts/readthedocs.org/user_builds/crowsetta/checkouts/latest/doc
/home/docs/checkouts/readthedocs.org/user_builds/crowsetta/envs/latest/lib/python3.10/site-packages/IPython/core/magics/osm.py:417: UserWarning: using dhist requires you to install the `pickleshare` library.
self.shell.db['dhist'] = compress_dhist(dhist)[-100:]
Our TextGrid files are in a sub-directory, ./data/r6pqa (the unique ID associated with the OSF project for the dataset).
We get a list of the TextGrid files from that subdirectory using pathlib.
To download these files and work with them locally, click the following links:
b06_concat_dtw.TextGrid
b11_concat_dtw.TextGrid
b35_concat_dtw.TextGrid
import pathlib
downloaded_dir = pathlib.Path('./data/r6paq/')
We make sure that our paths are sorted just in case
it matters
for our analysis.
textgrid_paths = sorted(downloaded_dir.glob('*.TextGrid'))
We inspect the first four paths just to check that we got what we expect.
textgrid_paths
[PosixPath('data/r6paq/b06_concat_dtw.TextGrid'),
PosixPath('data/r6paq/b11_concat_dtw.TextGrid'),
PosixPath('data/r6paq/b35_concat_dtw.TextGrid')]
Looks like it.
Each of the files corresponds to all the song from one individual bird. E.g., the file b06_concat_dtw.TextGrid contains annotations from the bird with ID “b06” and the file b011_concat_dtw.TextGrid contains annotations from the bird with ID “b11”.
We will need to do some book-keeping as we transform our data so we that we’re able to fit a model for each bird. Mainly this will involve using Python dictionaries where the bird’s ID is the key, and pandas DataFrames where we have the ID in a column.
But first let’s load all those annotations with crowsetta to get a sense of them.
import crowsetta
scribe = crowsetta.Transcriber(format='textgrid')
Notice below the argument keep_empty=True. The parameter keep_empty defaults to False, but we actually want to keep the unlabeled intervals between syllables, because they will help us divide the song up into bouts, as we will see below.
These are large files (that concatenate annotations across a day of song for each bird) so they take a while to load.
textgrids = {}
for textgrid_path in textgrid_paths:
bird_id = textgrid_path.name.split('_')[0] # bird ID will be first element from list after split
annot = scribe.from_file(textgrid_path, keep_empty=True)
textgrids[bird_id] = annot
2. Munge data#
Now we want to munge the data into the format we want to carry out our analysis.
Looking at descriptive statistics#
A good first thing to do with any new dataset is plot some descriptive statistics to get an overview of the data before we do anything with it. As an example of this, let’s look at the counts of the different classes of labels.
First we convert the TextGrid annotations to crowsetta.Annotations to make them easier to work with.
Here we use a dictionary comprehension to transform our textgrids dictionary to a dictionary of crowsetta.Annotations.
annots = {bird_id: textgrid.to_annot()
for bird_id, textgrid in textgrids.items()}
Now we count the distribution of labels. We loop over the dictionary values since for now we don’t care as much about the labels on a bird-by-bird basis (you’ll see why after we inspect them).
from collections import Counter
counts = Counter(
[lbl for annot in annots.values() for lbl in annot.seq.labels]
)
print(counts)
Counter({'': 53428, 'x': 20970, 'd': 4924, 'e': 4520, 'b': 3992, 'c': 3896, 'a': 3807, 'f': 2878, 'g': 2147, 'h': 1524, 'l': 1209, 'j': 1203, 'k': 1200, 'i': 1158})
We can see that we have many segments in our sequences that have the label 'x'.
These are squawky “intro” notes that are sometimes ommitted from models of song syntax.
In fact this is the most frequent label!
(The collection.Counter prints its key-value pairs in descending order of value by default.)
We will remove the squawky “intro” notes for this analysis (this is sometimes called “cleaning” data, although maybe you shouldn’t make your data feel like it’s dirty).
Dividing the annotations into song bouts#
We want to add states to our model for the beginning and ending of each song bout. To do that with the data we’re using here, we need to find the bouts within the annotations somehow, since each annotation file contains entire days of song.
One way to do this is to set a threshold on the duration of silent intervals, e.g. greater than 500 seconds, and use that threshold to break up annotations into bouts.
Before we do that, let’s eyeball the distribution of the silent intervals with empty string labels, to see what kind of threshold might make sense.
We start by converting the annotations to a pandas.DataFrame so we can easily munge and tidy the data. This is one advantage of moving from the Praat TextGrid format to the 'generic-seq' format built into crowsetta.
One approach would be to put all the annotations in a single DataFrame, adding a column named 'bird ID' that we’d use to split the DataFrame back up below (a sort of “long form” DataFrame). But we’ll stick with a dictionary to hopefully make the loops a bit easier to follow without inspecting the contents of DataFrames. Our new dictionary dfs will map each bird ID to a pandas.DataFrame made from its annotations.
dfs = {
bird_id: crowsetta.formats.seq.GenericSeq(annots=annot).to_df()
for bird_id, annot in annots.items()
}
for bird_id, df in dfs.items():
df['duration'] = df['offset_s'] - df['onset_s']
import matplotlib.pyplot as plt
fig, ax_arr = plt.subplots(2, 2, figsize=(10,8), dpi=150)
ax_arr = ax_arr.ravel()
for ax_ind, (bird_id, df) in enumerate(dfs.items()):
ax = ax_arr[ax_ind]
if ax_ind < 2:
bins = 100 # so we don't end up with one giant bin with most values in it
else:
bins = None # default is fine
df[df.label == ''].hist(column='duration', bins=bins, ax=ax)
ax.set_title(bird_id)
ax.set_xlabel('duration')
ax.set_xlim([0, 2])
rects = ax.patches
for rect in rects[3:18:2]:
height = rect.get_height()
ax.text(rect.get_x() + rect.get_width() / 2, height+0.01, f'{int(height)}',
ha='center', va='bottom', rotation=-45)
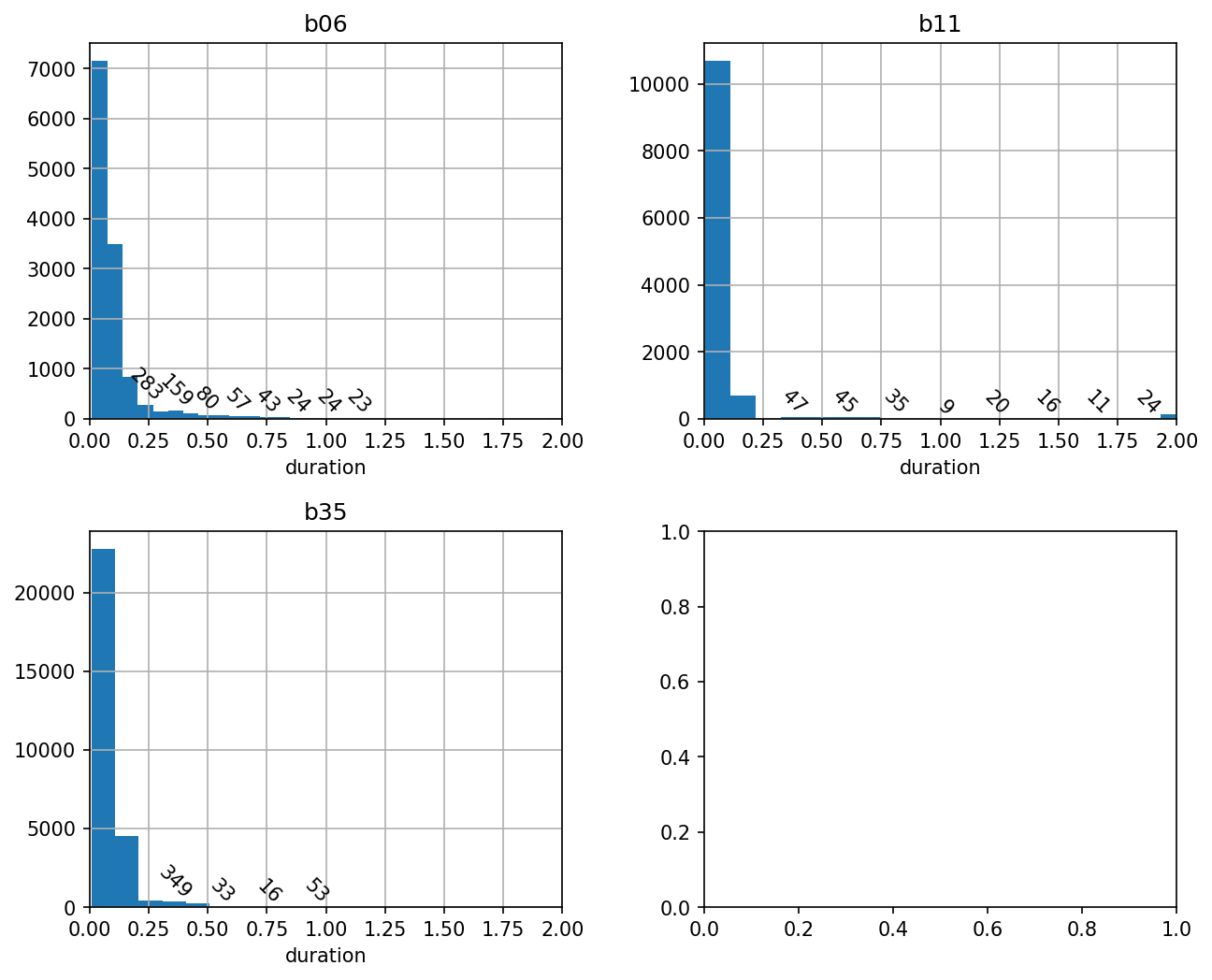
We can see that far and away the most frequent silent intervals are very brief, consistent with what is known about Bengalese finch song, where the silent gaps between syllables are 50-100 ms.
We can also see there’s a long tail of durations that are greater than 500 milliseconds.
To keep this vignette as simple as possible, let’s arbitrarily choose that threshold as “good enough”,
and then use it to split our annotations into bouts.
At the same time that we do this, we’ll remove all the other silent labels, which was our goal in the first place.
We continue to work with the pandas.DataFrame for convenience.
First we make a Boolean column that is True where our conditions hold of a segment with a silent label that has a duration greater than our threshold.
THRESHOLD = 0.5
for bird_id, df in dfs.items():
df['between_bouts'] = (df.duration > THRESHOLD) & (df.label == '')
Now we can find all the places that are True using numpy.nonzero, and then use those indices to split our pandas.DataFrame up into a list of smaller pandas.DataFrame, our bouts. To do the splitting, we use the numpy.array_split function, that can interface with pandas.DataFrames.
import numpy as np
bouts = {}
for bird_id, df in dfs.items():
inds = np.nonzero(df['between_bouts'].values)[0].tolist()
bouts_bird = np.array_split(df, inds)
bouts[bird_id] = bouts_bird
/home/docs/checkouts/readthedocs.org/user_builds/crowsetta/envs/latest/lib/python3.10/site-packages/numpy/core/fromnumeric.py:59: FutureWarning: 'DataFrame.swapaxes' is deprecated and will be removed in a future version. Please use 'DataFrame.transpose' instead.
return bound(*args, **kwds)
/home/docs/checkouts/readthedocs.org/user_builds/crowsetta/envs/latest/lib/python3.10/site-packages/numpy/core/fromnumeric.py:59: FutureWarning: 'DataFrame.swapaxes' is deprecated and will be removed in a future version. Please use 'DataFrame.transpose' instead.
return bound(*args, **kwds)
/home/docs/checkouts/readthedocs.org/user_builds/crowsetta/envs/latest/lib/python3.10/site-packages/numpy/core/fromnumeric.py:59: FutureWarning: 'DataFrame.swapaxes' is deprecated and will be removed in a future version. Please use 'DataFrame.transpose' instead.
return bound(*args, **kwds)
Let’s check bouts for one the birds and see how many that gave us.
We use the set of bouts from the last time through the for loop in the cell above, since it’s still hanging out in memory.
len(bouts_bird)
227
Seems like a reasonable number. What do the bouts look like?
Are they all more or less the same length?
bouts_sorted = sorted(bouts_bird, key=lambda bout: len(bout))
for bout in bouts_sorted[:25]:
print(len(bout))
2
16
22
24
24
30
30
30
30
34
40
40
40
42
42
48
48
48
52
52
56
56
62
62
64
Hmm, the first few look quite short. Let’s inspect one.
bouts_bird[1]
| label | onset_s | offset_s | notated_path | annot_path | sequence | annotation | duration | between_bouts | |
|---|---|---|---|---|---|---|---|---|---|
| 260 | 20.829 | 21.678 | None | data/r6paq/b35_concat_dtw.TextGrid | 0 | 0 | 0.849 | True | |
| 261 | x | 21.678 | 21.730 | None | data/r6paq/b35_concat_dtw.TextGrid | 0 | 0 | 0.052 | False |
| 262 | 21.730 | 21.818 | None | data/r6paq/b35_concat_dtw.TextGrid | 0 | 0 | 0.088 | False | |
| 263 | x | 21.818 | 21.904 | None | data/r6paq/b35_concat_dtw.TextGrid | 0 | 0 | 0.086 | False |
| 264 | 21.904 | 22.002 | None | data/r6paq/b35_concat_dtw.TextGrid | 0 | 0 | 0.098 | False | |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 537 | i | 45.523 | 45.580 | None | data/r6paq/b35_concat_dtw.TextGrid | 0 | 0 | 0.057 | False |
| 538 | 45.580 | 45.599 | None | data/r6paq/b35_concat_dtw.TextGrid | 0 | 0 | 0.019 | False | |
| 539 | j | 45.599 | 45.670 | None | data/r6paq/b35_concat_dtw.TextGrid | 0 | 0 | 0.071 | False |
| 540 | 45.670 | 45.707 | None | data/r6paq/b35_concat_dtw.TextGrid | 0 | 0 | 0.037 | False | |
| 541 | k | 45.707 | 45.850 | None | data/r6paq/b35_concat_dtw.TextGrid | 0 | 0 | 0.143 | False |
282 rows × 9 columns
We can see the one above begins with has many of the silent intervals or the ‘x’ class that we don’t care about. We can filter these intervals out and then inspect our bouts again.
How about a longer DataFrame with more rows in it, though?
bouts_sorted[-1]
| label | onset_s | offset_s | notated_path | annot_path | sequence | annotation | duration | between_bouts | |
|---|---|---|---|---|---|---|---|---|---|
| 49042 | 4314.169 | 4314.670 | None | data/r6paq/b35_concat_dtw.TextGrid | 0 | 0 | 0.501 | True | |
| 49043 | x | 4314.670 | 4314.697 | None | data/r6paq/b35_concat_dtw.TextGrid | 0 | 0 | 0.027 | False |
| 49044 | 4314.697 | 4314.900 | None | data/r6paq/b35_concat_dtw.TextGrid | 0 | 0 | 0.203 | False | |
| 49045 | x | 4314.900 | 4314.962 | None | data/r6paq/b35_concat_dtw.TextGrid | 0 | 0 | 0.062 | False |
| 49046 | 4314.962 | 4315.110 | None | data/r6paq/b35_concat_dtw.TextGrid | 0 | 0 | 0.148 | False | |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 51491 | e | 4518.538 | 4518.647 | None | data/r6paq/b35_concat_dtw.TextGrid | 0 | 0 | 0.109 | False |
| 51492 | 4518.647 | 4518.694 | None | data/r6paq/b35_concat_dtw.TextGrid | 0 | 0 | 0.047 | False | |
| 51493 | f | 4518.694 | 4518.814 | None | data/r6paq/b35_concat_dtw.TextGrid | 0 | 0 | 0.120 | False |
| 51494 | 4518.814 | 4518.859 | None | data/r6paq/b35_concat_dtw.TextGrid | 0 | 0 | 0.045 | False | |
| 51495 | x | 4518.859 | 4518.933 | None | data/r6paq/b35_concat_dtw.TextGrid | 0 | 0 | 0.074 | False |
2454 rows × 9 columns
This looks more like a real bout of song, with labels corresponding to the syllables we are interested in. But we still also have silent periods and our ‘x’ segments as well.
Let’s definitely filter all the rows out that are unlabeled, as we set out to do, and the rows where the label is ‘x’ too.
bouts_filtered = {}
for bird_id, bouts_bird in bouts.items():
bouts_bird = [
bout[(bout.label != '') & (bout.label != 'x').copy()]
for bout in bouts_bird
]
bouts_filtered[bird_id] = bouts_bird
After we do that we can inspect our DataFrames again and see if they’re more like sequences of syllables.
for bout in bouts_filtered['b35'][:10]:
print(len(bout))
83
77
46
109
52
24
30
23
45
20
Not surprisingly they are shorter. There may be some empty “bouts” that we need to discard.
What do the bouts look like after filtering?
bouts_filtered['b35'][3].head()
| label | onset_s | offset_s | notated_path | annot_path | sequence | annotation | duration | between_bouts | |
|---|---|---|---|---|---|---|---|---|---|
| 737 | a | 64.726 | 64.822 | None | data/r6paq/b35_concat_dtw.TextGrid | 0 | 0 | 0.096 | False |
| 739 | b | 64.896 | 65.019 | None | data/r6paq/b35_concat_dtw.TextGrid | 0 | 0 | 0.123 | False |
| 741 | c | 65.043 | 65.192 | None | data/r6paq/b35_concat_dtw.TextGrid | 0 | 0 | 0.149 | False |
| 743 | d | 65.220 | 65.369 | None | data/r6paq/b35_concat_dtw.TextGrid | 0 | 0 | 0.149 | False |
| 747 | f | 65.650 | 65.762 | None | data/r6paq/b35_concat_dtw.TextGrid | 0 | 0 | 0.112 | False |
Yes, this actually looks like a sequence of syllables.
Let’s filter out the empty “bouts”.
bouts_filtered = {
bird_id: [bout for bout in bouts if len(bout) > 0]
for bird_id, bouts in bouts_filtered.items()
}
Now finally we’ll make new annotated sequences that we can pass to the functions we wrote above.
annots_munged = {}
for bird_id, bouts in bouts_filtered.items():
annots= []
for bout in bouts:
bout = bout.reset_index() # so we can get row '0' below
onsets_s, offsets_s, labels = bout.onset_s.values, bout.offset_s.values, bout.label.values
seq = crowsetta.Sequence.from_keyword(onsets_s=onsets_s, offsets_s=offsets_s, labels=labels)
annot_path = bout.loc[0, 'annot_path'] # will be the same for all rows
annot = crowsetta.Annotation(annot_path=annot_path, seq=seq)
annots.append(annot)
annots_munged[bird_id] = annots
So how many bouts did we end up with? We check for one bird.
len(annots_munged['b35'])
223
This is a bit low for four days of song, but good enough number to fit a model for our purposes here.
We inspect the Sequence of the first Annotation just to further confirm that it worked.
annots_munged['b35'][0].seq.segments[:10]
(Segment(label='b', onset_s=1.263, offset_s=1.376, onset_sample=None, offset_sample=None),
Segment(label='c', onset_s=1.399, offset_s=1.547, onset_sample=None, offset_sample=None),
Segment(label='d', onset_s=1.572, offset_s=1.716, onset_sample=None, offset_sample=None),
Segment(label='e', onset_s=1.941, offset_s=2.052, onset_sample=None, offset_sample=None),
Segment(label='f', onset_s=2.099, offset_s=2.219, onset_sample=None, offset_sample=None),
Segment(label='g', onset_s=2.249, offset_s=2.351, onset_sample=None, offset_sample=None),
Segment(label='h', onset_s=2.391, offset_s=2.481, onset_sample=None, offset_sample=None),
Segment(label='i', onset_s=2.51, offset_s=2.565, onset_sample=None, offset_sample=None),
Segment(label='j', onset_s=2.582, offset_s=2.655, onset_sample=None, offset_sample=None),
Segment(label='k', onset_s=2.684, offset_s=2.814, onset_sample=None, offset_sample=None))
Yes, this looks like what we saw in the DataFrame above, converted to annotations.
We have now transformed the annotations into the format we want for our analysis.
This is a point in our analysis pipeline where we go from one form of the data for another, so it’s probably a good place to save what we’ve done.
We do this by making a new set of 'generic-seq’ annotations, and then saving to a csv file.
for bird_id, annots_list in annots_munged.items():
annots_munged_generic = crowsetta.formats.seq.GenericSeq(annots=annots_list)
annots_munged_generic.to_file(
f'./data/tachibana-morita-2021-munged-for-markov-bird-{bird_id}.csv'
)
4. Compute transition matrix#
Okay, let’s finally build a Markov model!
All we want now is the labels attributes from our sequence-like annotations; we will throw away the rest of the information for this step.
We then pass the labels into the pipeline we made with our functions above.
models = {}
for bird_id, annots in annots_munged.items():
labels = [annot.seq.labels for annot in annots]
counts, matrix, states = transmat_from_labels(labels)
models[bird_id] = (counts, matrix, states)
Lastly we visualize the transition matrices as heatmaps.
import seaborn as sns
sns.set_context('notebook')
sns.set(font_scale=0.25)
fig, ax_arr = plt.subplots(2, 2, dpi=300)
for ind, (bird_id, (counts, matrix, states)) in enumerate(models.items()):
ax = ax_arr.ravel()[ind]
g = sns.heatmap(matrix,
xticklabels=states, yticklabels=states[1:],
annot=True, fmt=".2f",
vmin=0., vmax=1.,
ax=ax);
ax.set_title(bird_id)
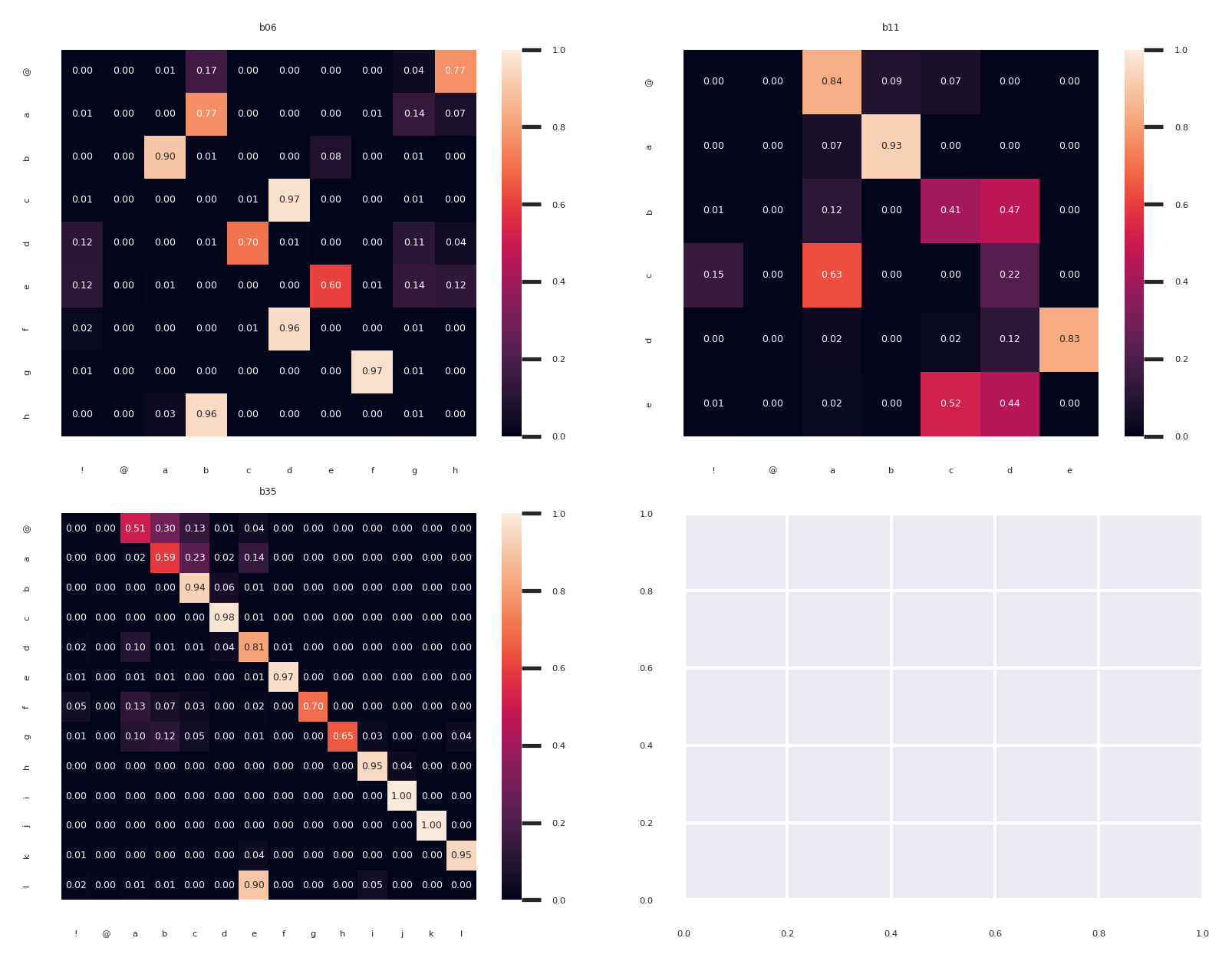
Comparing across birds we can see that some individuals have a very linear song with the most frequent transitions on the diagonal (like b35 in the bottom right), while others have more variable transitions (b17 in the bottom left).
Now you’ve seen a detailed example of using crowsetta to access and transform annotations in a specific format in order to fit a simple model of behavior.
